Dr. Tsung-Lin Liu

Dr. Tsung-Lin Liu
Position:Professor
Group:Biomedical technology
Research Interests:Bioinformatics, Next-generation Sequencing Data Analysis, Metagenomics
E-mail:tsunglin@mail.ncku.edu.tw
Room:89904
Room Tel:+886-6-2757575#58223
Laboratory Tel:+886-6-2757575#58234#911

|
School |
Department |
Country |
Degree |
Period |
|
Ohio State University |
Department of Physics |
USA |
Ph.D. |
2000~2006 |
|
National Taiwan University |
Department of Physics |
TW |
B.S. |
1993~1997 |

|
Institute |
Position |
Period |
|
Department of Biotechnology and Bioindustry Sciences, National Cheng-Kung University (NCKU) |
Professor |
2023~now |
|
Department of Biotechnology and Bioindustry Sciences, National Cheng-Kung University (NCKU) |
Associate Professor |
2014~2023 |
|
Institute of Bioinformatics and Biosignal Transduction, National Cheng-Kung University (NCKU) |
Assistant Professor |
2010~2014 |
|
Biodiversity Research Center, Academia Sinica, Taiwan |
Postdoctoral Fellow |
2008~2010 |
|
Neuroscience Research Institute, UCSB, U.S.A. |
Postdoctoral Fellow |
2006~2008 |
|
in Physics, OSU, U.S.A. |
Ph.D. |
2000~2006 |
|
Institute of Atomic and Molecular Sciences, Academia Sinica, Taiwan |
Research Assistant |
1999~2000 |
Bioinformatics is an interdisciplinary study that applies informatics techniques to analyze biological data with accuracy and efficiency. With the introduction of next-generation sequencing (NGS), the field of bioinformatics thrives to handle the huge amount of data of different natures. Among bioinformatics topics, we focus on metagenomics and immune repertoire analysis. Metagenomics applies NGS to explore microbial communities in many environments, including human body sites, soils, and water. We collaborate with clinical doctors in studying association between human microbiome and health, e.g., between nasal microbiota and childhood asthma, between oral microbiota and gargling agents, between gut microbiota and intestinal bowel disease. Our works have pointed out important microbes involved in several human diseases. Immune repertoire depicts the diversity of T cells via tracking their unique T-cell receptor (TCR) sequences. Unique TCR sequences are results of the so-called VDJ recombination. Non-recombined TCR sequences are usually considered as garbage as they do not code for proteins. However, we found that non-recombined TCR sequences can be a biomarker of T-cell lymphoma. We are exploring more novel functions of non-recombined TCR sequences.
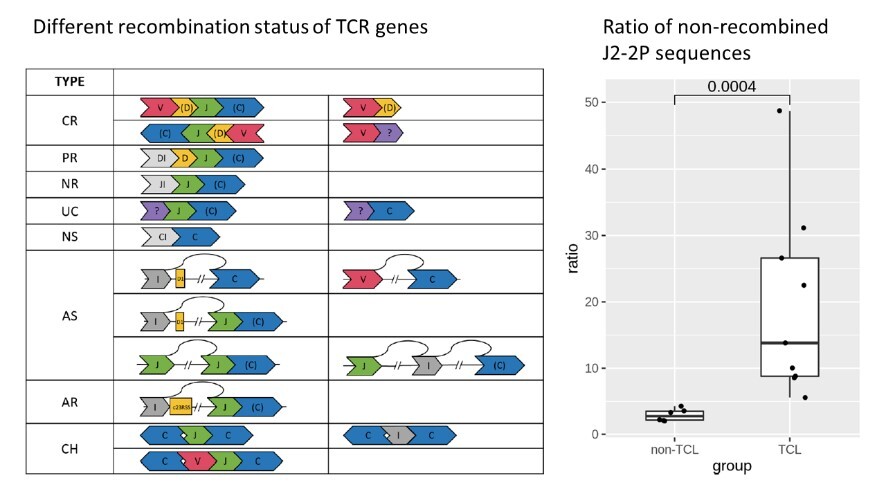 |
Clonality assessment, which can detect neoplastic T cells by identifying the uniquely recombined T-cell receptor (TCR) genes, provides important support in the diagnosis of T-cell lymphoma (TCL). BIOMED-2 is the gold standard clonality assay and has proven to be effective in European TCL patients. However, we failed to prove its sensitivity in Taiwanese TCL patients, especially based on the TCRb gene. Interestingly, we found a higher percentage (>81%) of non-recombined TCRb sequences in a TCL patient with a negative BIOMED-2 test. This suggests a new TCR target for enhancing TCL diagnosis. To further explore the hypothesis, we proposed a cost-effective digital PCR assay that quantifies the relative abundance of non-recombined TCRb sequences containing a J2-2P~J2-3 segment. With the digital PCR assay, bone marrow specimens from TCL patients (n=9) showed a positive outcome (i.e., the relative abundance of the J2-2P~J2-3 sequences ≧5%), whereas non-TCL patients (n=6) gave a negative result. As five of nine TCL patients had a negative BIOMED-2 test result, the J2-2P~J2-3 sequences may improve TCL detection. This is the first report showing the capability of characterizing non-recombined TCR sequences as a supplementary strategy for the BIOMED-2 clonality test. |
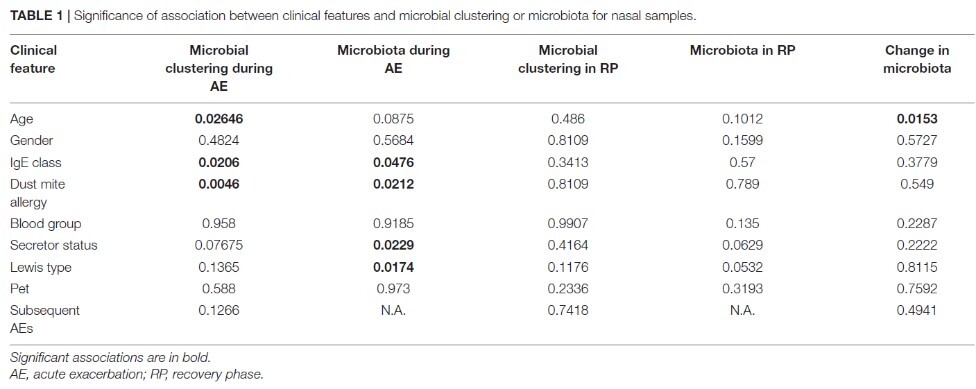 |
Airway and gut microbiota are important in asthma pathogenesis. Although several studies have revealed distinct microbiota in asthmatic airways at baseline compared to healthy controls, limited studies compared microbiota during acute exacerbation (AE) and in the recovery phase (RP) in the same asthmatic children. We aim to investigate association between microbiota and asthma status in children and explore their relationship with clinical features of asthma. Using 16S rRNA sequencing, we found that most nasal microbiota were dominated by only one or two of six bacterial genera. The domination was associated with mite allergy and patient age only during AE but not in the RP. When moving into RP, the relative abundance of Staphylococcus increased while that of Moraxella decreased. Throat and stool microbiota were not associated with most of the clinical features. Interestingly, stool microbiota during AE was associated with ABO blood type and stool microbiota in the RP was associated with frequency of the subsequent exacerbations. In summary, the association between nasal microbiota and mite allergy only during AE suggests an altered local immunity and its interplay with nasal microbes. Our work provides a basis for studying microbes, and prevention or therapeutic strategy in childhood asthma, especially during AE. |
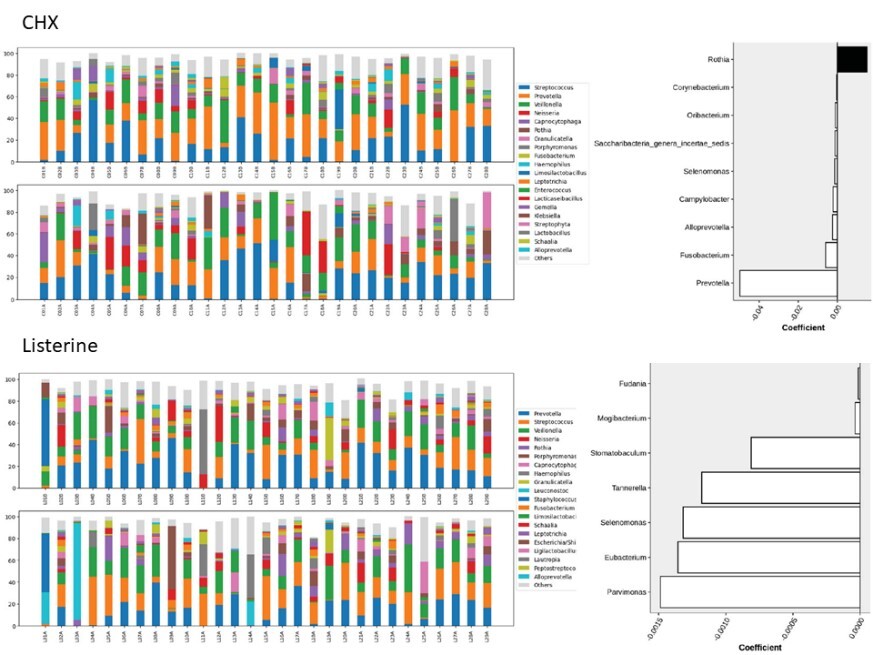 |
Chlorhexidine (CHX) and essential oil containing mouthwashes like Listerine® can improve oral hygiene via suppressing oral microbes. In hospitalized patients, CHX mouthwash reduces the incidence of ventilator-associated pneumonia. However, CHX use was also associated with increased mortality, which might be related to nitrate-reducing bacteria. Currently, no study determines oral bacteria targeted by essential oils mouthwash in hospitalized patients using a metagenomic approach, which is the aim of this work. Paired analysis revealed eight bacterial genera (including Prevotella, Fusobacterium, and Selenomonas) with a decreased relative abundance, while Rothia increased after gargling the CHX mouthwash. After gargling Listerine, seven genera (including Parvimonas, Eubacterium, and Selenomonas) showed a decreased relative abundance, and the magnitudes were smaller compared to the CHX group. Fewer bacteria targeted by Listerine were reported to be nitrate-reducing compared to the CHX mouthwash. In conclusion, short-term gargling of the CHX mouthwash and Listerine altered oral microbiota in our hospitalized patients. The bacterial genera targeted by the CHX mouthwash and Listerine were largely different and the magnitudes of changes were smaller using Listerine. Functional alterations of gargling CHX and Listerine were also different. These findings can be considered for managing oral hygiene of hospitalized patients. |


.svg.png)
